TriPepSVM
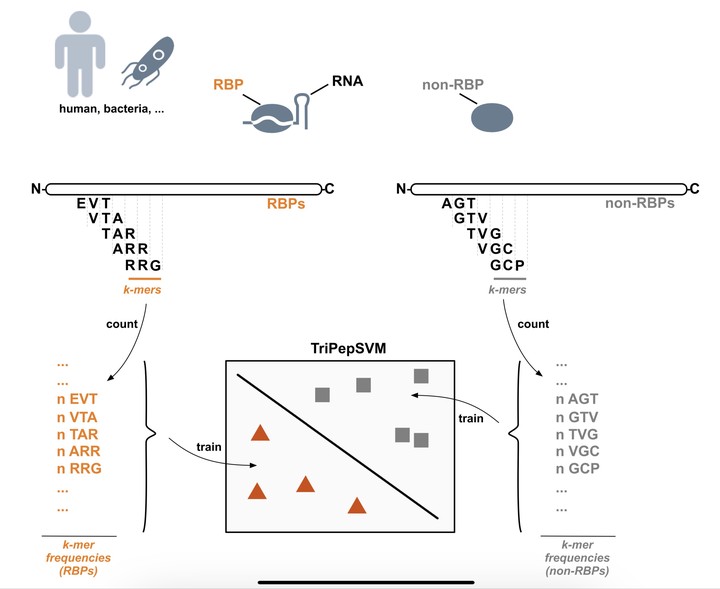
RNA-binding proteins (RBPs) are well studied in eukaryotes such as human, mouse or yeast. In bacteria however, the landscape of cellular RBPs was much harder to study, mainly due to technical reasons. In a collaborative project with the lab of Annalisa Marsico (Computational RNA Biology Group, Helmholtz Centrum München) , we developed a machine-learning-based approach to predict RBPs: TriPepSVM. We use a support vector machine which is trained on short amino acid motifs to screen bacterial and eukaryotic proteomes for RNA-binders.
We could successfully apply TriPepSVM to E. coli and Salmonella Typhimurium. In Salmonella, we predicted 66 novel RBPs from which we tested some in the lab, thereby confirming that 3 out of 4 predictions are correct.
The work has been published and can be found here .